LitGene
Gene-Disease-Drug Association.
Introduction
LitGENE is an interpretable transformer-based model that integrates textual information and gene ontologies through contrastive learning to refine gene representations. The approach combines gene annotation with gene description to predict gene functions and attributes. This is achieved by integrating contrastive learning within a large language model.
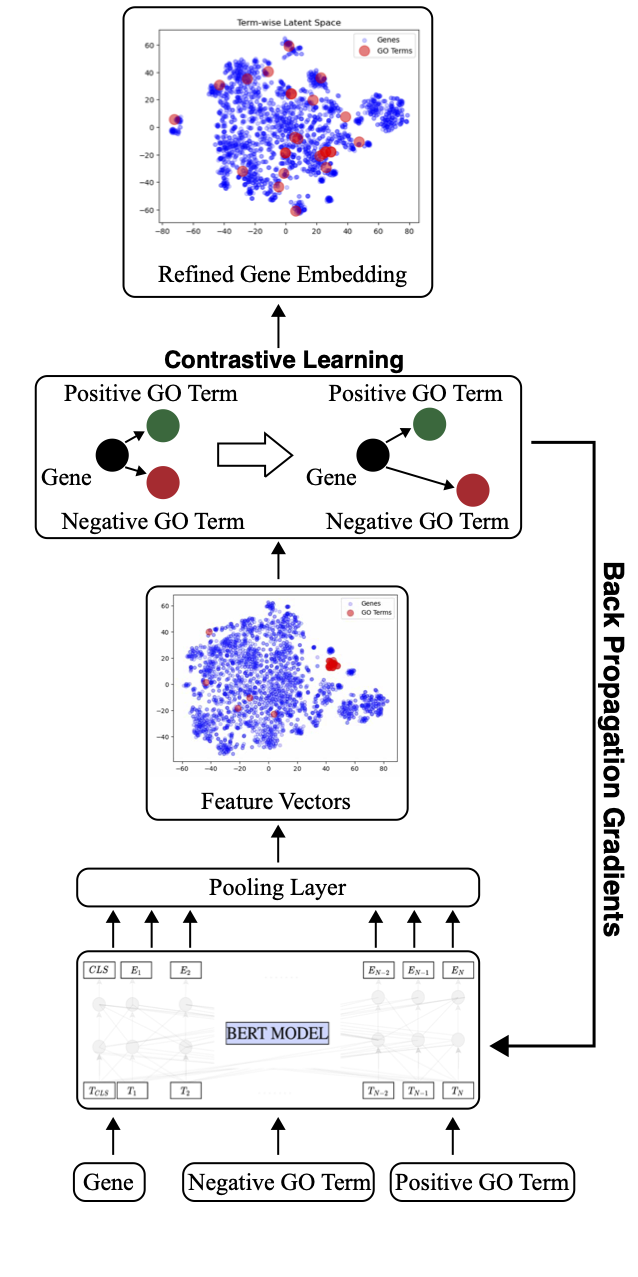
Figure 1: Overview of LitGENE Model
Inputs
The website accepts descriptions of biomedical entities, such as genes/proteins, pathways, diseases, or drugs. Researchers and clinicians can input scientific abstracts (in the prompt window) related to these entities and receive predictions of related biological entities.
Outputs
LitGENE provides predictions of biomedical entities related to the input prompt.
-
For Example:
Disease Input: Outputs include predicted risk genes associated with the disease and drugs that target these genes, potentially modulating disease severity.
Additional Outputs
- Similarity Scores: Indicate the strength of the predicted relationships.
- Word Importance: Reports which words in the input are most important in determining the predicted relationships.
- Citations: Provides references from the published literature supporting the predicted relationships. References are inferred by LitGENE from among 100,000 PubMed entries.